Distinguished lecture (Hong Kong Baptist University. Dept. of Computer Science)

Will AI Replace Professors?
2025 | 53 mins
Geometric Deep Graph Learning: A New Perspective on Graph Foundation Model
2024 | 63 mins
Big AI for Small Devices
2024 | 55 mins
The Hidden Truths of Principal Component Analysis
2024 | 83 mins
Artificial Intelligence (AI): Past, Present and Future
2024 | 91 mins
Data Valuation in Federated Learning
2024 | 73 mins
Biomedicine in the Age of Generative AI
2024 | 63 mins
Understanding Non-Verbal Communication: Decoding Facial Expressions and Gestures for Seamless Human-Robot Interaction
2024 | 71 mins
The Impact of ChatGPT on Artificial Intelligence Research and Social Development
2023 | 65 mins
Cardinality Estimation of Approximate Substring Queries using Deep Learning
2023 | 67 mins
Towards Production Federated Learning Systems
2023 | 76 mins
Bioinformatics Challenges in Complete Genomics and Metagenomics
2023 | 67 mins
Improving Object Detection from Repeated Traversals of the Same Route
2023 | 59 mins
Querying Geo-Textual Data Using Keywords – A Retrospective
2023 | 72 mins
People Analytics: Using Digital Exhaust from the Web to Leverage Network Insights in the Algorithmically Infused Workplace
2023 | 73 mins
The State of Representing and Solving Games
2023 | 83 mins
Towards Sustainable Edge and IoT: An Integral View from the Energy Perspective
2023 | 67 mins
Commodifying Data Exploration
2023 | 55 mins
Towards Interdisciplinary Education in the Metaverse
2023 | 73 mins
Past, Present, and Future of Feature Extraction
2023 | 59 mins
The Data Landscape: Trends and Directions
2022 | 61 mins
Fairness in Algorithmic Systems: A Reality or a Fantasy?
2022 | 75 mins
Personal Identity, Artificial Intelligence, and Biometric Entropy
2022 | 63 mins
Challenges in Machine Learning Research
2022 | 72 mins
Machine Learning for Big Spatial Data and Applications
2022 | 64 mins
A Vision towards Pervasive Edge Computing
2021 | 81 mins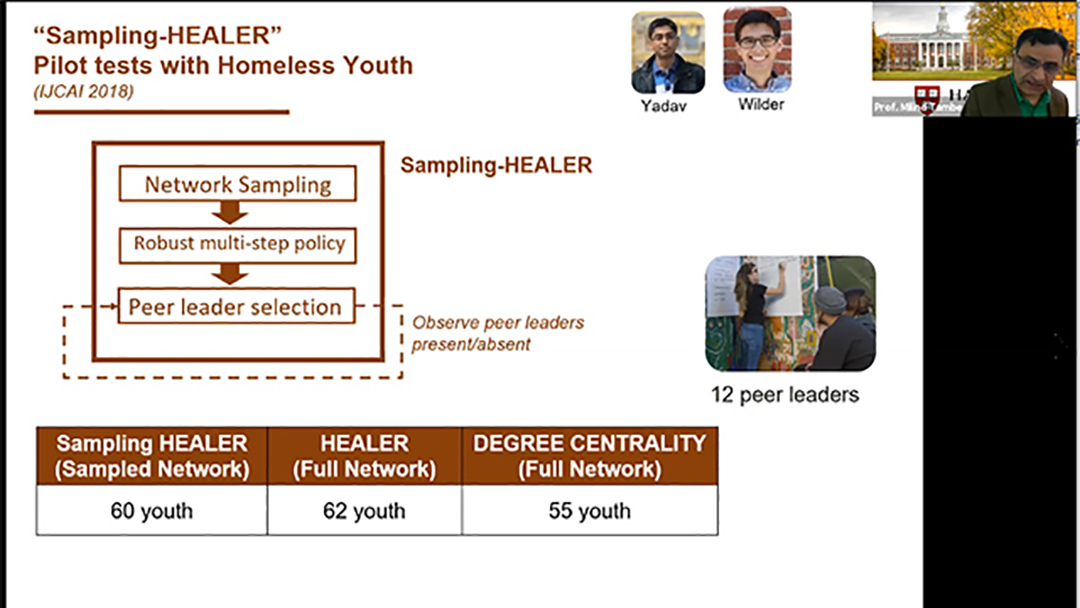
AI for social impact: Results from deployments for public health and conservation
2021 | 63 mins
Blockchain in Action: Business Models, Application Examples, and Strategies
2019 | 60 mins
Blockchains Untangled: Public, Private, Smart Contracts, Applications, Issues
2019 | 96 mins
From the Edge of Biometrics: What's Next?
2018 | 77 mins
Deep Representations, Adversarial Learning and Domain Adaptation for Some Computer Vision Problems
2018 | 69 mins
Developmental Brain Storm Optimization Algorithms
2018 | 75 mins
Non-iterative Learning Methods
2018 | 54 minsData-driven Evolutionary Optimization – Integrating Evolutionary Computation, Machine Learning and Data Sciences
2018 | 73 mins
Enabling High-Quality and Efficient Vehicle Routing using Massive Trajectory Data
2018 | 64 mins
Entropic Analysis of Time Evolving Networks
2018 | 75 mins
How to Achieve Data Analytics and Artificial Intelligence in X
2018 | 77 mins
Recommender Systems: Beyond Machine Learning
2018 | 80 mins
Personalized Transfer Learning
2017 | 74 mins
Face Clustering at Scale
2017 | 72 mins
Multimedia over Future Internet: Challenges and Opportunities
2017 | 62 mins
Adversarial Signal Processing and The Hypothesis Testing Game
2017 | 68 mins
Special Sensing System for Computer Vision
2017 | 72 mins
50+ Years of Fuzzy Sets: Machine Intelligence and Data to Knowledge
2017 | 81 mins
Building a Biomedical Research Digital Commons
2017 | 71 mins
My Smartphone Knows What You Print: Exploring Smartphone-based Side-channel Attacks Against 3D Printers
2016 | 57 mins
Visual Domain Adaptation
2017 | 81 mins
Making Sense of Spatial Trajectories
2016 | 80 mins
New Correlation Filter Designs and Applications
2016 | 79 mins
Artificial Intelligence: Key to Chinese Manufacturing 2025
2016 | 75 mins
Who Is There? Face Recognition Applications
2015 | 70 mins
Beauty and the Computer
2015 | 59 mins
Big Data for Everyone
2015 | 82 mins
Biometric Recognition: Technology for Human Recognition
2015 | 82 mins
Is Computer Vision Pattern Recognition by a Different Name?
2015 | 67 mins
Optimization in Machine Learning
2015 | 78 mins
New Challenges in Multi-robot Task Allocation
2015 | 68 mins
Some Mysteries of Multiplication, and How to Generate Random Factored Integers
2015 | 62 mins
The HDL Lola and its Translation to Verilog
2015 | 59 mins
Computers and Computing in the Early Years
2015 | 83 mins
The Oberon System on a Field-Programmable Gate Array (FPGA)
2015 | 74 mins
Urban Computing : Using Big Data to Solve Urban Challenges
2014 | 70 mins
Keyword-Based Spatial Web Querying
2014 | 70 mins
Preserving Individual and Institutional Privacy in Distributed Regression Models
2014 | 68 mins
The Spiral of Theodorus, Numerical Analysis, and Special Functions
2014 | 61 mins
The Evolution of Probabilistic Models and Uncertainty Analysis in Computer Vision Research
2014 | 72 mins
Towards Connecting Big Data with Many People
2014 | 66 mins
Mapping the Next Generation Human Health on a Global Scale : Eight E-health Mega-themes and Trends
2013 | 104 mins
Self-Learning Control of Nonlinear Systems based on Iterative Adaptive Dynamic Programming Approach
2013 | 57 mins
Mobile Cloud and Crowd Computing, Communications and Sensing
2013 | 78 mins
A 50-Year Personal Odyssey in Computer Science
2012 | 84 mins
Fusing Multiple Sources of Conflicting Information
2011 | 83 mins
Dimensionality Reduction for Real-Time Autonomous Systems
2011 | 64 mins
Machine Intelligence, F-granulation and Generalized Rough Sets: Uncertainty Analysis in Pattern Recognition and Mining
2011 | 95 mins
Discovery of Cyclic GMP and Nitric Oxide cell signaling and their role in drug development
2010 | 79 mins
Product Form Solutions for Stochastic Networks : Discovery or Invention?
2010 | 74 mins
How Precise Documentation Allows Information Hiding to Reduce Software Complexity and Increase Its Agility
2009 | 90 mins
Computers and Music
2013 | 22 mins
Learning to Simplify Sonography during and after the Ultrasound Examination
2025 | 12 mins
Liver Cancer Prediction: From Single Time-Point to Multimodality
2025 | 15 mins
Bridging the Interpretability Gap for Medical AI Models Using a Generative Explanation Approach
2025 | 25 mins
Exploration in Medical AI
2025 | 19 mins
Intelligent Neuromodulation with Edge Computing
2025 | 25 mins
Leveraging 30 Years of Clinical Data and AI Solutions to Advance Healthcare Research, Innovation, and Service in Hong Kong
2025 | 23 mins
AI for Population Health
2025 | 46 mins
Application of AI in Liver Diseases
2025 | 29 mins







